This page was produced as an assignment for Genetics 677, an undergraduate course at UW-Madison.
Future Directions
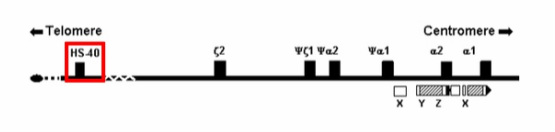
Modified from Galanello and Cao [1].
Something that struck my interest while researching the primary literature on the HBA genes was the HS-40 upstream regulatory region. The presence of this region is just as critical for α-globin synthesis as the HBA genes themselves [1]. Therefore, I would propose that future research be conducted to observe the effects of manipulating the transcription factors that bind to this region.
Using String, I identified GATA-1 and NF-E2 as proteins known to associate with HBA1 and HBA2 [2]. A literature search confirmed both of these proteins are transcription factors that bind to sites in the HS-40 region [1].
Using String, I identified GATA-1 and NF-E2 as proteins known to associate with HBA1 and HBA2 [2]. A literature search confirmed both of these proteins are transcription factors that bind to sites in the HS-40 region [1].
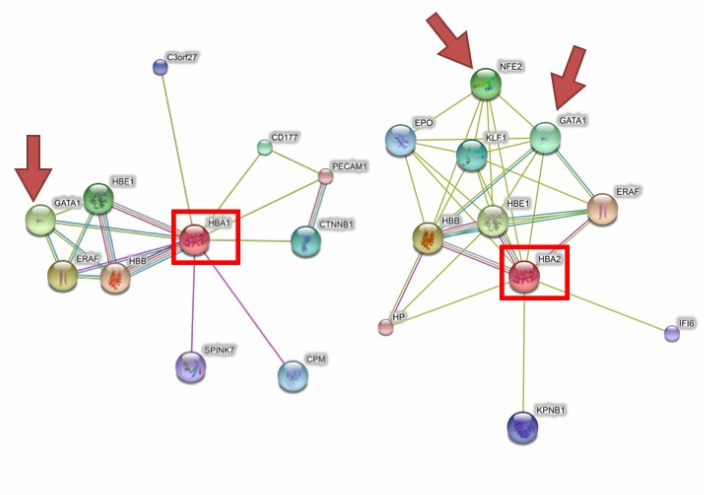
Interaction webs for HBA1 and HBA2 modified from String [2], with GATA-1 and NF-E2 indicated.
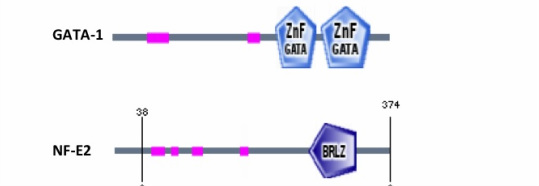
Modified from SMART [3].
A motif search on SMART showed that GATA-1 and NF-E2 both contain DNA binding domains [3]. GATA-1 contains two zinc finger domains that bind to GATA sequences in HS-40, while NF-E2 has a leucine zipper binding domain.
I would like to explore the relationship between the expression of these two transcription factors and the HBA genes in a mouse model. The mouse homologue of HBA1, Hba-a1 is highly conserved (See Homologeny and Phylogeny). The knockout mouse strain Hba-a1(tm1Led) (See RNAi and Model Organisms) has an intact Hba-a2 gene. Mice also have an regulatory region upstream of their α-globin genes, called HS-26. This region contains most of the binding sites present in the human HS-40 region, including those for GATA-1 and NF-E2 [4].
In order to explore the effects of changes in GATA-1 and NF-E2 on the mouse Hba genes, I would propose a microarray analysis. First, I would design a transgenic mouse line that overexpressed GATA-1 and NF-E2, and cross these mice to an Hba-a1 knockout strain. The resulting progeny would have overexpressed GATA-1 and NF-E2 in the absence of Hba-a1. I would then perform a microarray analysis to compare the expression of the intact Hba-a2 gene in these double transgenic mice to wild-type and each of the transgenes alone.
I would like to explore the relationship between the expression of these two transcription factors and the HBA genes in a mouse model. The mouse homologue of HBA1, Hba-a1 is highly conserved (See Homologeny and Phylogeny). The knockout mouse strain Hba-a1(tm1Led) (See RNAi and Model Organisms) has an intact Hba-a2 gene. Mice also have an regulatory region upstream of their α-globin genes, called HS-26. This region contains most of the binding sites present in the human HS-40 region, including those for GATA-1 and NF-E2 [4].
In order to explore the effects of changes in GATA-1 and NF-E2 on the mouse Hba genes, I would propose a microarray analysis. First, I would design a transgenic mouse line that overexpressed GATA-1 and NF-E2, and cross these mice to an Hba-a1 knockout strain. The resulting progeny would have overexpressed GATA-1 and NF-E2 in the absence of Hba-a1. I would then perform a microarray analysis to compare the expression of the intact Hba-a2 gene in these double transgenic mice to wild-type and each of the transgenes alone.
Expected Results
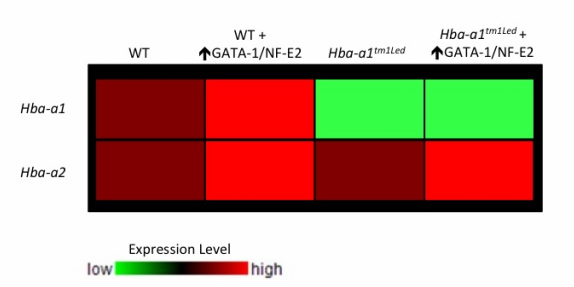
I hypothesize that my proposed experiments would yield results similar to those seen here. The data, portrayed as a microarray heat map, would likely show that the GATA-1/NF-E2 overexpression mice would increase expression of both Hba-a1 and Hba-a2 in a wild-type background. I predict that I would also see up-regulated expression of the intact Hba-a2 gene. My hope would be that this increased expression of Hba-a2 would be sufficient to compensate for the missing Hba-a1 gene and potentially rescue the α-thalassemia symptoms in the knockout mice. If successful in mice, the idea of artificially up-regulating GATA-1 and NF-E2 in human α-thalassemia patients could be a potential method for gene therapy approaches to treat the disease.
References
- Alpha-Thalassemia. GeneReviews [Internet]. Galanello, R., Cao, A. Seattle (WA): University of Washington, Seattle; 1993-. 2005 Nov 01 [last updated 2008 July 15]. (NCBI Bookshelf)
- The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, Jensen LJ, von Mering C. Nucleic Acids Res. 2011 Jan;39(Database issue):D561-8. Epub 2010 Nov 2. (PubMed)
- SMART, a simple modular architecture research tool: Identification of signaling domains. Schultz, J., Milpetz, F., Bork, P. & Ponting, C.P. PNAS 1998; 95: 5857-5864. (PubMed)
- Properties of the mouse alpha-globin HS-26: relationship to HS-40, the major enhancer of human alpha-globin gene expression. Bouhassira EE, Kielman MF, Gilman J, Fabry MF, Suzuka S, Leone O, Gikas E, Bernini LF, Nagel RL. Am J Hematol. 1997 Jan;54(1):30-9. (PubMed)
