This page was produced as an assignment for Genetics 677, an undergraduate course at UW-Madison.
Microarray
Using NCBI's Gene Expression Omnibus (GEO) database [1], I searched for microarray data on HBA1. There was very little information on HBA1, but there some expression data for HBA2 was available. None of the data sets were from work on α-thalassemia, but rather the expression of HBA2 was observed in a large set of genes in a study relating to other conditions. One study by Raghavachari, et al. found that HBA2 was up-regulated in sickle cell disease, while the beta and delta hemoglobin genes were down-regulated. Interestingly, this study was the second to find that globin genes were highly expressed in platelets, not just erythrocytes. [2]
Though limited microarray data relating to α-thalassemia was available, microarrays may be used to diagnose the disease. In 2004, researchers at Shenzhen Yishengtang Biological Products developed microarrays with deletion-specific probes designed to hybridize to PCR products amplified from DNA of patients carrying the three most common deletional alleles of α-thalassemia (see figure below). They found that their detection method was highly reproducible in identifying these three alleles. Their paper also mentions that microarray studies of α-thalassemia can be difficult due to high homology in the α-globin gene cluster, which complicates PCR primer design and oligonucleotide probe selection. [3] These obstacles may be one of the reasons that so little microarray data relating to α-thalassemia is available.
Though limited microarray data relating to α-thalassemia was available, microarrays may be used to diagnose the disease. In 2004, researchers at Shenzhen Yishengtang Biological Products developed microarrays with deletion-specific probes designed to hybridize to PCR products amplified from DNA of patients carrying the three most common deletional alleles of α-thalassemia (see figure below). They found that their detection method was highly reproducible in identifying these three alleles. Their paper also mentions that microarray studies of α-thalassemia can be difficult due to high homology in the α-globin gene cluster, which complicates PCR primer design and oligonucleotide probe selection. [3] These obstacles may be one of the reasons that so little microarray data relating to α-thalassemia is available.
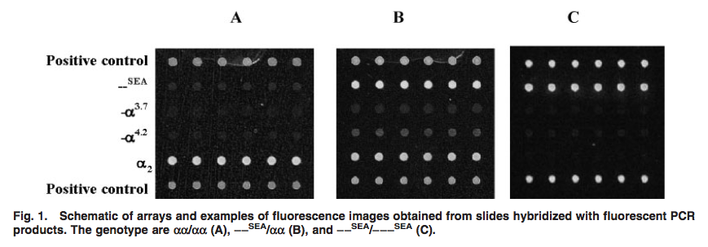
Figure 1 from the paper by Zesong, Ruijun, and Wen, illustrating their diagnostic microarrays. SEA = Southeast Asian allele. The other alleles, α3.6 and α4.2, are deletions of different sizes. [3].
References
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002 Jan 1;30(1):207-10. (PubMed)
- Amplified expression profiling of platelet transcriptome reveals changes in arginine metabolic pathways in patients with sickle cell disease. Raghavachari N, Xu X, Harris A, Villagra J, Logun C, Barb J, Solomon MA, Suffredini AF, Danner RL, Kato G, Munson PJ, Morris SM Jr, Gladwin MT. Circulation. 2007 Mar 27;115(12):1551-62. Epub 2007 Mar 12. (PubMed)
- Rapid detection of deletional alpha-thalassemia by an oligonucleotide microarray. Zesong L, Ruijun G, Wen Z. Am J Hematol. 2005 Dec;80(4):306-8. (PubMed)
