This page was produced as an assignment for Genetics 677, an undergraduate course at UW-Madison.
Motifs
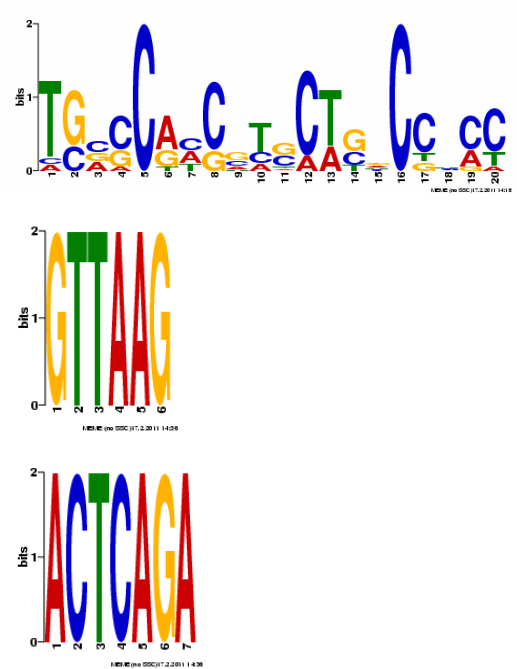
These DNA motifs were generated by MEME. Nucleotide sequence motifs are often widespread in a gene sequence, and may have biological significance. Here, motifs 2 and 3 have very consistent sequences where they are found throughout the gene. Additionally, motif 3 is found in both the 5' and 3' termini of the gene. [1] Though motifs 2 and 3 are very distinct in the HBA1 sequence, a BLAST search did not produce any homologues.
Using another site, Motif, I was able to find 7 DNA motifs: an EGF1-like domain, an integrins beta chain cysteine-rich domain, a C-terminal cystine knot, an anaphylatoxin domain, a VWFC domain, 2Fe-2S ferredoxins, iron-sulfur binding region, and a mammalian defensins motif [2]. By clicking on the links to the domains, you can see (via PROsite), that each of these domains are found in proteins in a vast range of species.
References
- Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Timothy L. Bailey and Charles Elkan. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, pp. 28-36, AAAI Press, Menlo Park, California, 1994. (PubMed) http://meme.sdsc.edu/meme4_6_0/intro.html
- A generalized profile syntax for biomolecular sequences motifs and its function in automatic sequence interpretation. Bucher P., Bairoch A. ISMB-94; Proceedings 2nd International Conference on Intelligent Systems for Molecular Biology. (Altman R., Brutlag D., Karp P., Lathrop R., Searls D., Eds.), pp53-61, AAAIPress, Menlo Park, 1994. (PubMed)
