This page was produced as an assignment for Genetics 677, an undergraduate course at UW-Madison.
Homology and Alignments
Protein Alignment
Generated using T-COFFEE [1,2,3]. Alignment of Homo sapiens HBA1 protein sequence with homologous sequences in Bos taurus (cattle), Mus musculus (mouse), Rattus norvegicus (rat), and Gallus gallus (chicken). The sequences are shown to be highly homologous by this alignment. Additionally, close HBA1 homologues are found only in higher vertebrates. This is not surprising, as HBA1 serves an oxygen-carrying role that makes its presence most necessary in organisms with highly developed vascular systems.
Another Alignment
This alignment was generated using ClustalW [1,2,4]. The alignment is similar to the one generated by T-COFFEE. However, I preferred the format of the T-COFFEE alignment, as the color coding gives you a quick visual assessment of homology.
Generated using T-COFFEE [1,2,3]. Alignment of Homo sapiens HBA1 protein sequence with homologous sequences in Bos taurus (cattle), Mus musculus (mouse), Rattus norvegicus (rat), and Gallus gallus (chicken). The sequences are shown to be highly homologous by this alignment. Additionally, close HBA1 homologues are found only in higher vertebrates. This is not surprising, as HBA1 serves an oxygen-carrying role that makes its presence most necessary in organisms with highly developed vascular systems.
Another Alignment
This alignment was generated using ClustalW [1,2,4]. The alignment is similar to the one generated by T-COFFEE. However, I preferred the format of the T-COFFEE alignment, as the color coding gives you a quick visual assessment of homology.
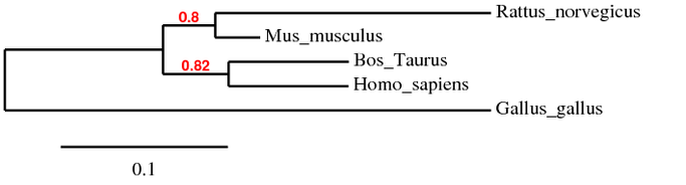
Phylogenetic Tree
This phylogeny was generated using TreeDyn [5], and showing the conservation of HBA1 in animals with complex vasculature.
References
- BLAST-EXPLORER helps you building datasets for phylogenetic analysis. Dereeper A., Audic S., Claverie J.M., Blanc G.BMC Evol Biol. 2010 Jan 12;10:8. (PubMed)
- O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Dereeper A.*, Guignon V.*, Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.F., Guindon S., Lefort V., Lescot M., Claverie J.M., Gascuel Nucleic Acids Res. 2008 Jul 1;36(Web Server issue):W465-9. Epub 2008 Apr 19. (PubMed) *: joint first authors
- T-Coffee: A novel method for fast and accurate multiple sequence alignment. Notredame C., Higgins DG., Heringa J. J Mol Biol. 2000, Sep 8;302(1):205-17. (PubMed)
- CLUSTAL W: improving the sensitivity of progressivemultiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Thompson J.D., Higgins D.G., Gibson T.J. Nucleic Acids Res. 1994, Nov 11;22(22):4673-80. (PubMed)
- TreeDyn: towards dynamic graphics and annotations for analyses of trees. Chevenet F., Brun C., Banuls A.L., Jacq B. and R. Christen. BMC Bioinformatics 2006, 7:439. (PubMed)